
neoSwitch™ enables binder discovery and protein production in the same host under control of a chemical trigger in the growth media, eliminating weeks of subcloning and protein expression in a secondary host which is normally required to obtain soluble protein for characterization (SPR, ELISA etc). neoSwitch comes pre-transformed with high diversity libraries for ‘plug and play’ yeast display workflows.

Validation of neoSwitch
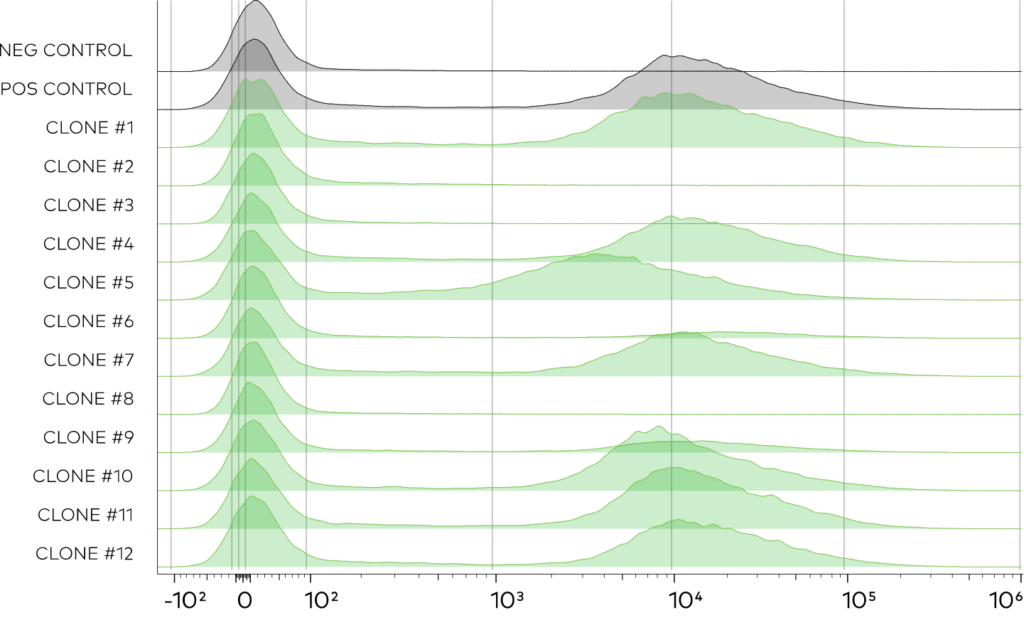
After transformation,12 random clones were selected to measure surface display by flow cytometry with an anti-Myc secondary. In agreement with bulk-measurements indicating ~75% of the library was functional, most clones were properly displayed and expressed at levels comparable to a validated control VHH. It was concluded that this library is sufficient for antigen specific VHH discovery campaigns.
To discover binders to CD5, the neoVHH™ library was subjected to 2 rounds of positive MACS selections, 3 rounds of negative MACS selections, and 2 rounds of positive FACS selections. From a naive library of 1.5e10 yeast cells, 2e7 cells were sorted onto agar plates. On-target and off-target binding was measured for 90 colonies, of which 35 had unique CDR3 sequences, highlighting the quality of the neoVHH library. 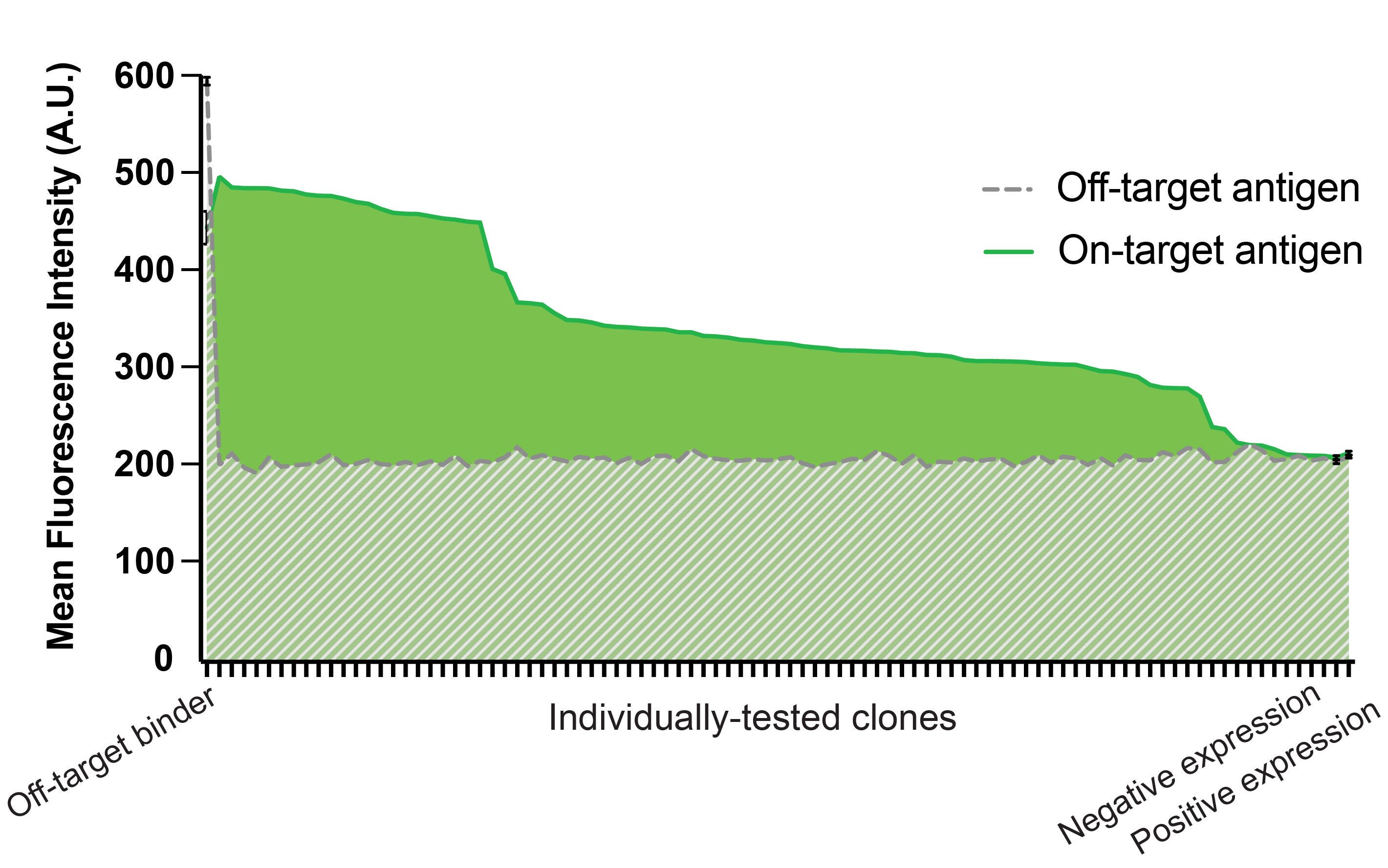
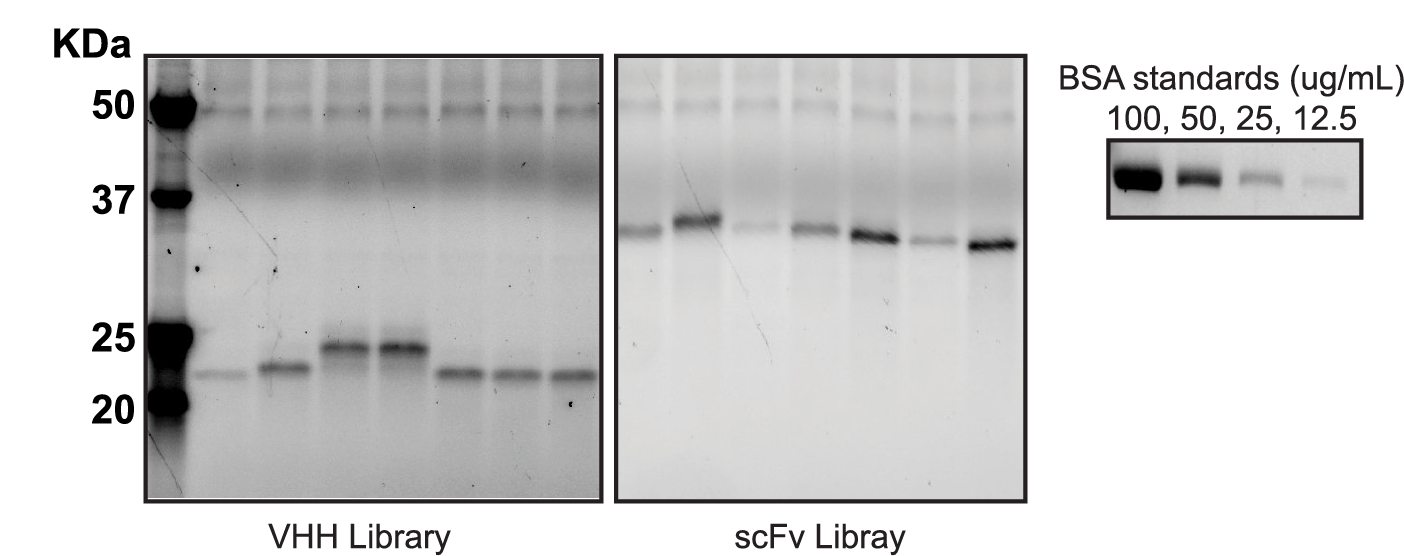
Secretion yields of VHH and scFv were determined by stain free PAGE-gel of supernatant. VHH and scFv libraries grown for 48 hours at 30C in 96-well plates in secretion mode. VHH clones had measurable secretion in the range of 10-50ug/mL and scFv clones had measurable secretion in the range of 50-100ug/mL. Secretion yields were quantified by densitometry compared to a standard curve of known concentration.
Production of purified binders is useful to enable screening assays including epitope binning, stability studies, and cell-based activity assays. To validate affinity purification of secreted VHH from neoSwitchTM, we grew a 200mL culture and AKTA purified His-tagged VHH from the supernatant by Ni-NTA affinity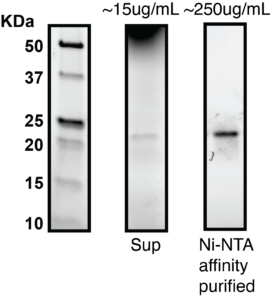
Iterative campaigns to generate diversity from top hits
Product Features
Library Specifications and TAT
| Neo Designed Synthetic VHH Library | Llama VHH Library | Ships Immediately |
| Customer-Defined Library | Custom Mutational Schema | Subject to Library Build Time |
| Customer Existing Starting Material | e.g. cDNA from immunization, pre-existing library | Subject to Library Build Time |
Synthetic Naive Library for Antibody Discovery
- Controlled mutagenesis library biased for CDRs to create functional binders
- CDR3 is a mixture of 3 different lengths
- Sequence liabilities (e.g. cysteine and methionine) are removed