neoDisplay™ offers turnkey access to yeast display, a highly sensitive method of binder discovery leveraging fluorescence activated cell sorting (FACS) for isolation of developable, specific, and high affinity binders. neoDisplay™ comes pre-transformed with high diversity libraries for ‘plug and play’ yeast display workflows. Perform affinity maturation and humanization campaigns more rapidly by combining libraries delivered in neoDisplay™ with Neo’s 3-day sequencing and rapid variant library construction.
Library Specifications and TAT
| Neo Designed Synthetic VHH Library | Llama VHH Library | Ships Immediately |
| Customer-Defined Library | Custom Mutational Schema | Subject to Library Build Time |
| Customer Existing Starting Material | e.g. cDNA from immunization, pre-existing library | Subject to Library Build Time |
neoDisplay
Synthetic VHH Library
- Controlled mutagenesis library biased for CDRs to create functional binders
- CDR3 is a mixture of 3 different lengths
- Sequence liabilities (e.g. cysteine and methionine) are removed
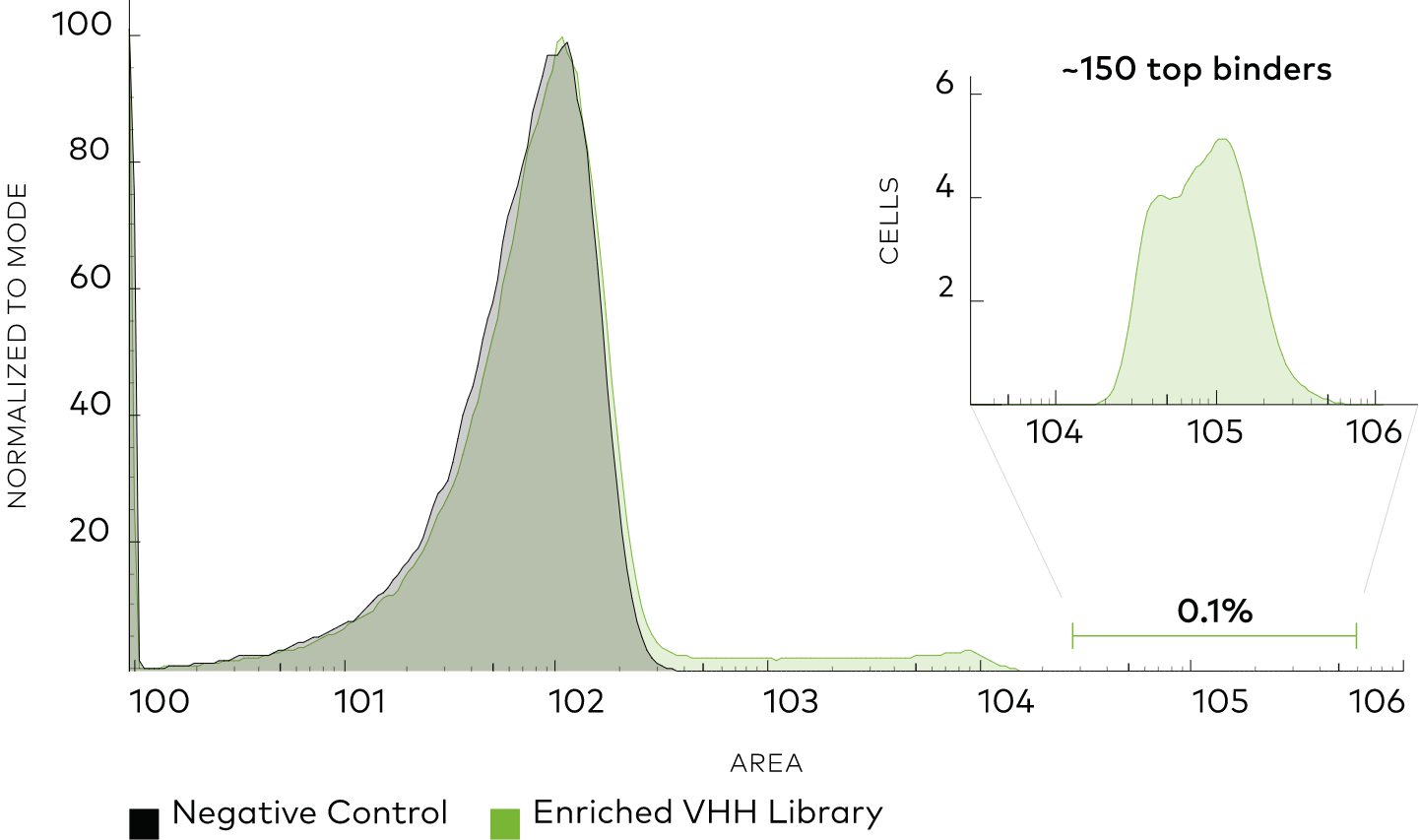
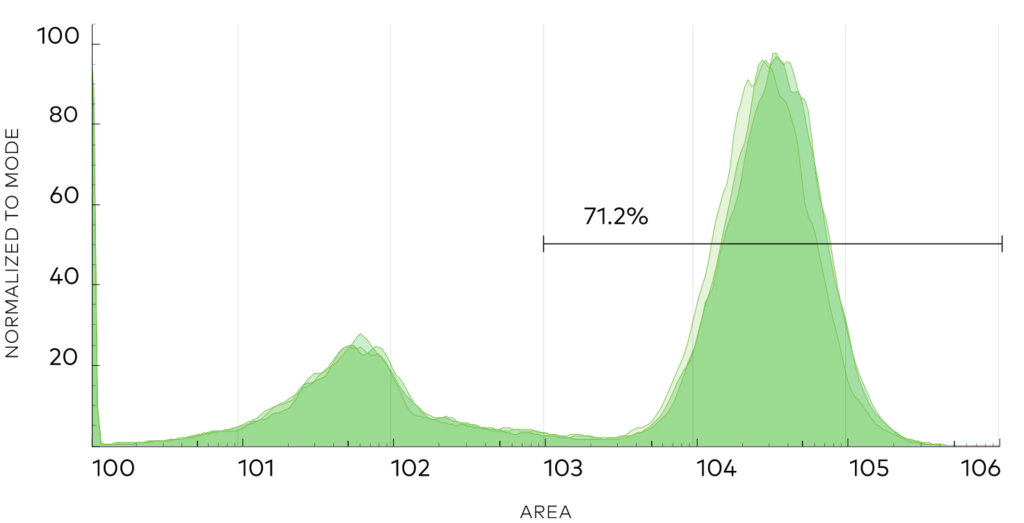
Using neoDisplay™ transformed with Neo’s Synthetic VHH library, we performed a discovery campaign to identify binders to PD1. We performed 2 rounds of negative selection to remove non-specific binding and 3 rounds of positive selection against PD1. We isolated 96 single cells from the positive sorting gate and identified 33 unique binder sequences (defined as at least 2 differences in CDR3 sequence).